Stomach Scaffold¶
The current stomach scaffold is 3D Stomach 1 built from class MeshType_3d_stomach1.
The human variant is shown in Fig. 79.
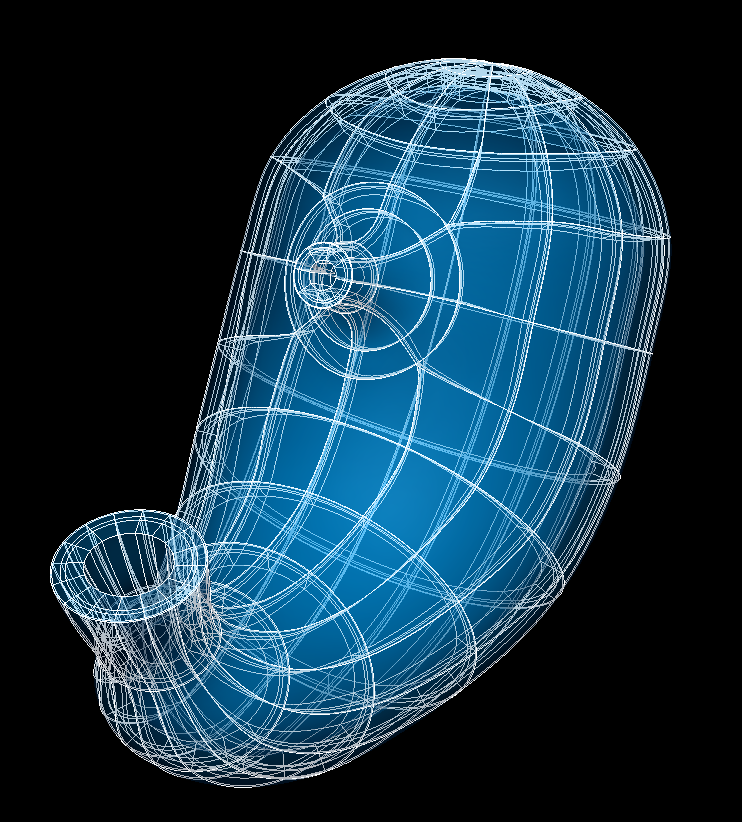
Fig. 79 Human stomach scaffold.¶
The stomach scaffold is a 3-D volumetric model of the stomach representing the fundus, corpus, cardia, antrum, pylorus, and a short distal section of the esophagus as well as a short proximal section of the duodenum.
Variants¶
The stomach scaffold is provided with parameter sets for the following four species, which differ in shape:
Human
Mouse
Pig
Rat
These variants’ geometry and annotations are best viewed in the Scaffold Creator tool in the ABI Mapping Tools.
On the web, the latest published generic stomach scaffold variants can be viewed on the
SPARC Portal by searching for stomach, filtering for anatomical models, selecting a
variant and viewing the scaffold in its Gallery tab or via the Organ Scaffolds help article.
The stomach scaffold script generates the scaffold mesh and geometry from an idealization of their shapes. The mesh is derived from ellipsoid and cubic functions based on a one dimensional central path with side axes controlling lateral dimensions. The parameters were carefully tuned for each species, and it is not recommended that these be edited.
Instructions for editing the central path are given with the ABI Mapping Tools Scaffold Creator documentation. Note that the D2 and D3 derivatives control the side dimensions, and derivatives D12 and D13 control the rate of change of these along the central path. If editing, use the Interactive Functions to Smooth derivatives, Make side derivatives normal and Smooth side cross derivatives to make these as smooth as required.
The rat stomach scaffold is parameterized with average data from segmentation of Micro-CT image data of 11 animals performed at the Powley laboratory using Neurolucida (MBF Bioscience), while the human, mouse, and pig stomach scaffolds are parameterized with literature data to represent the anatomy accurately.
The mucosa, submucosa, circular muscle, longitudinal muscle and serosa layers of the stomach are fully represented on
the scaffold when Number of elements through wall is set to 4. Alternatively, the entire stomach wall can be
represented as a single layer by setting Number of elements through wall to 1.
[A special Material parameter set is provided to allow new species’ parameters to be developed from the material
coordinates definition (see below).]
Coordinates¶
The stomach scaffold defines the geometric, split and material coordinates.
The geometric coordinates field gives an approximate, idealized representation of the stomach shape for the species,
which is intended to be fitted to actual data for a specimen.
The split coordinates follows the geometric field but provides disconnected and duplicated derivatives for nodes on
the margin between the dorsal and ventral stomach (the boundary where stomach specimens are usually dissected into
halves for imaging purposes). This field is intended for fitting data obtained from the dorsal or ventral stomach.
The material coordinates field stomach coordinates defines a highly idealized coordinate system to give permanent
locations for embedding structures in the stomach. It is defined by a capsule-shaped structure with an inlet cylindrical
tube representing the esophagus and another outlet tube representing the duodenum. This can be viewed by
visualising this field in the Display tab of Scaffold Creator or by switching to the special Material
parameter set.
The stomach scaffold supports limited refinement/resampling by checking Refine (set parameter to true) with chosen
Refine number of elements parameters. Be aware that only the coordinates field is currently defined on the refined
mesh (but annotations are transferred).
Annotations¶
Important anatomical regions of the stomach are defined by groups of elements (or faces, edges and nodes/points) and annotated with standard term names and identifiers from a controlled vocabulary.
Annotated 3-dimensional volume regions are defined by groups of 3-D elements including (using only one of the items separated by slash /):
body of stomach
cardia of stomach
circular/longitudinal muscle layer of stomach
dorsal stomach
duodenum
esophagogastric junction
esophagus
esophagus mucosa
esophagus smooth muscle circular/longitudinal layer
fundus of stomach
mucosa of stomach
pyloric antrum
pyloric canal
stomach
submucosa of esophagus
submucosa of stomach
ventral stomach
Terms for volume regions such as the above are not to be used for digitized contours! They are used for applying different material properties in models and the strain/curvature penalty (stiffness) parameters in fitting.
Annotated 2-dimensional surface regions are defined for matching annotated contours digitized from medical images
including (where surface is the outside boundary on the meshes and using only one of the items separated by slash
/):
circular-longitudinal muscle interface of dorsal stomach/stomach/ventral stomach
forestomach-glandular stomach junction (for mouse, rat or if Limiting ridge is checked)
gastroduodenal junction
greater/lesser curvature of stomach
luminal surface of body of stomach
luminal surface of cardia of stomach
luminal surface of duodenum
luminal surface of esophagus
luminal surface of fundus of stomach
luminal surface of pyloric antrum
luminal surface of pyloric canal
luminal surface of stomach
serosa of body of stomach
serosa of cardia of stomach
serosa of duodenum
serosa of esophagus
serosa of fundus of stomach
serosa of pyloric antrum
serosa of pyloric canal
serosa of stomach
Annotated 1-dimensional line regions are defined for matching annotated contours digitized from medical images including (using only one of the items separated by slash /):
circular-longitudinal muscle interface of body of stomach along the gastric-omentum attachment
circular-longitudinal muscle interface of esophagus along the cut margin
circular-longitudinal muscle interface of the first segment of the duodenum along the gastric-omentum attachment
circular-longitudinal muscle interface of fundus of stomach along the greater curvature
circular-longitudinal muscle interface of gastroduodenal junction
circular-longitudinal muscle interface of pyloric antrum along the greater/lesser curvature
circular-longitudinal muscle interface of pyloric canal along the greater/lesser curvature
limiting ridge on circular-longitudinal muscle interface/luminal surface/serosa (only in mouse, rat or if Limiting ridge is checked)
Several fiducial marker points are defined on the stomach scaffold, of which the followings are potentially usable when digitizing:
body-antrum junction along the greater curvature on circular-longitudinal muscle interface/luminal surface/serosa
distal point of lower esophageal sphincter serosa on the greater/lesser curvature of stomach
esophagogastric junction along the greater curvature on circular-longitudinal muscle interface/luminal surface/serosa
esophagogastric junction along the lesser curvature on circular-longitudinal muscle interface/luminal surface/serosa
fundus-body junction along the greater curvature on circular-longitudinal muscle interface/luminal surface/serosa (only in human, pig or if Limiting ridge is unchecked)
gastroduodenal junction along the greater curvature on circular-longitudinal muscle interface/luminal surface/serosa
gastroduodenal junction along the lesser curvature on circular-longitudinal muscle interface/luminal surface/serosa
limiting ridge at the greater curvature on serosa/the circular-longitudinal muscle interface/luminal surface (only in mouse, rat or if Limiting ridge is checked)